Studying back-to-back microbiome results
I sampled my gut microbiome three times over the month of May: once on May 1st and again on May 28th and May 29th.
Interestingly, the May 29th sample was taken after ingesting 250mg of Reducose the day before, giving me a hint at how Reducose may affect my microbiome.
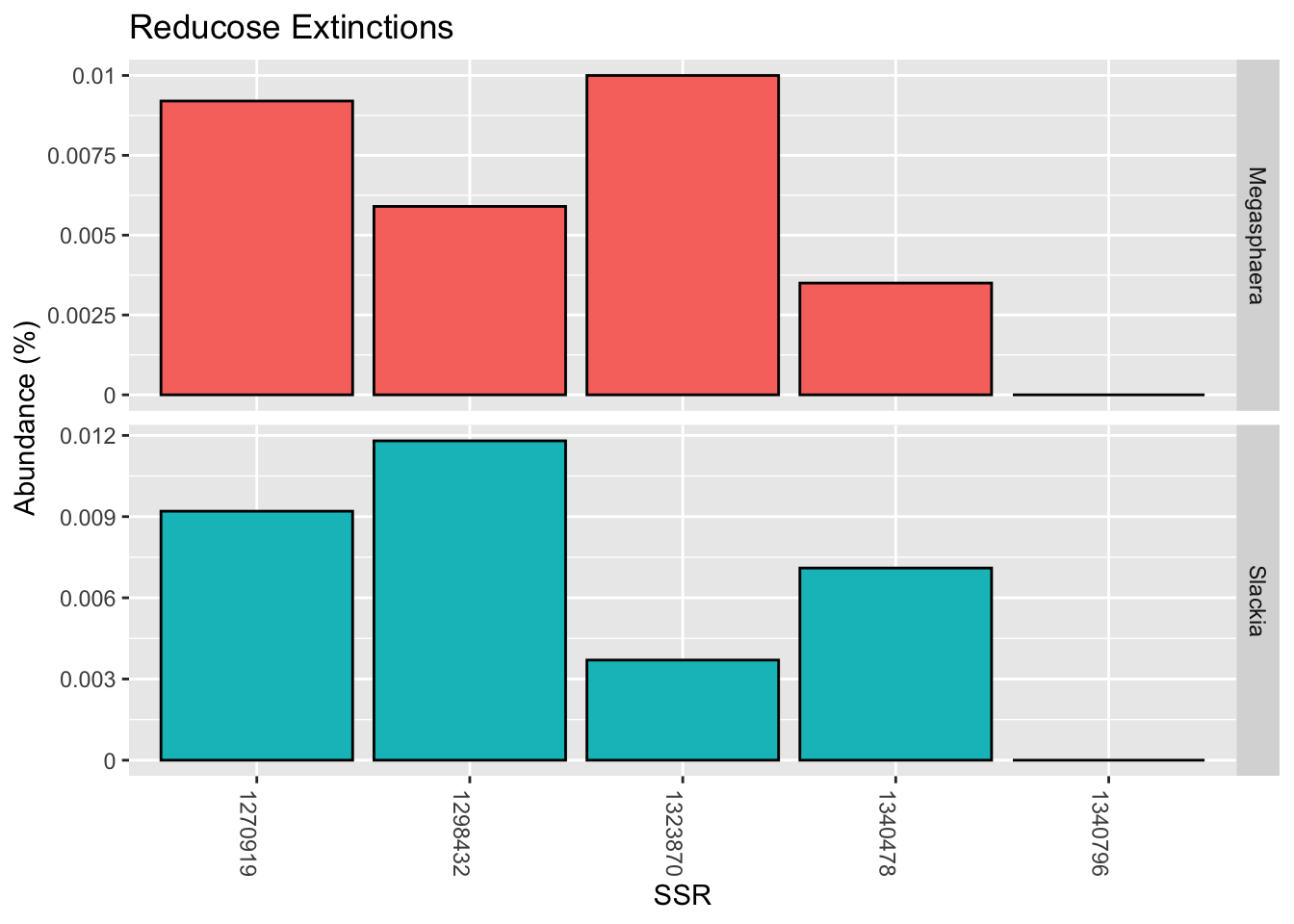
Here are the unique taxa found in the first sample (before Reducose) but not the second (genus level):
| Unique | % |
|---|---|
| Caldicoprobacter | 0.0023 |
| Enterobacter | 0.0119 |
| Fastidiosipila | 0.0059 |
| Megasphaera | 0.0035 |
| Murdochiella | 0.0023 |
| Prevotella | 0.0083 |
| Slackia | 0.0071 |
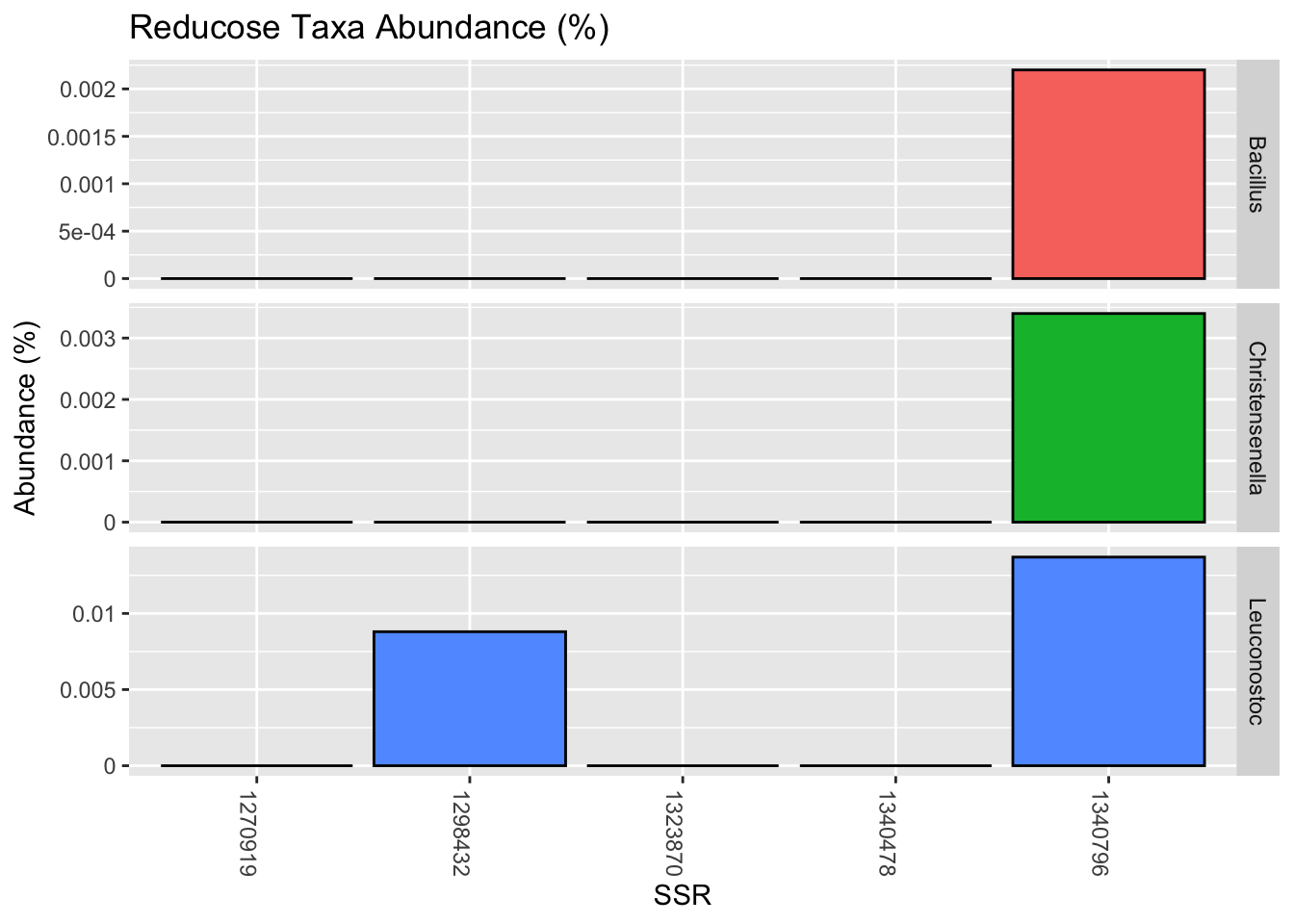
and here are those that appeared suddenly a day after ingesting Reducose:
| Extinct | % |
|---|---|
| Anaerofustis | 0.0045 |
| Bacillus | 0.0022 |
| Brevibacterium | 0.0068 |
| Christensenella | 0.0034 |
| Corynebacterium | 0.0091 |
| Eisenbergiella | 0.0034 |
| Leuconostoc | 0.0137 |
| Peptoniphilus | 0.0022 |
| Planomicrobium | 0.0022 |
and a heatmap:
Finally, let’s see which new microbes showed up between the beginning of the month and before taking the pre-Reducose sample:
| Unique | % |
|---|---|
| Anaerococcus | 0.0138 |
| Anaerofustis | 0.0037 |
| Brevibacterium | 0.0326 |
| Coprobacillus | 1.9770 |
| Corynebacterium | 0.1093 |
| Eisenbergiella | 0.6699 |
| Facklamia | 0.0025 |
| Hespellia | 0.0439 |
| Oligella | 0.0100 |
| Peptoniphilus | 0.0075 |
| Planomicrobium | 0.0025 |
| Porphyromonas | 0.0201 |
| Pseudoclavibacter | 0.0163 |
| Rhodococcus | 0.0062 |
| Staphylococcus | 0.0037 |
| Succiniclasticum | 0.0062 |
and the unique microbes that showed up during the same period:
| Extinct | % |
|---|---|
| Aggregatibacter | 0.0023 |
| Butyrivibrio | 0.0107 |
| Granulicatella | 0.0023 |
| Haemophilus | 0.0071 |
Interestingly, three of the microbes that were present before disappeared after the Reducose. Although this is a very tentative analysis based on one person, with only three samples, without additional evidence it points to some microbes I’d want to watch more seriously next time.